Databases in R – Simple, Fast, and Secure


For professionals working with large datasets in their daily routines, databases are an invaluable tool. As an electronic management system, databases are designed to efficiently handle large volumes of data while maintaining consistency and integrity. Additionally, within a company, a database ensures that all employees have access to a unified and up-to-date dataset. Any changes made to the database are immediately available to all stakeholders, which is especially beneficial when data is processed automatically by computer systems.
Mastering Databases in R with These Packages
R provides several packages for working with databases, making it easy to establish connections from within R and integrate databases into the data science workflow. For example, using the DBI and RMySQL packages, we can seamlessly connect to a test database (test_db) and explore the flights table, which contains information about departures from New York airports in 2013.
# Pakete laden
library(DBI) # Funktionen zum Umgang mit Datenbanken
library(RMySQL) # MySQL Treiber
library(dplyr) # Für %>%
# Konnektor-objekt erzeugen
con %
dbGetQuery("SELECT month, day, carrier, origin, dest, air_time
FROM flights LIMIT 3")
# month day carrier origin dest air_time
# 1 1 1 UA EWR IAH 227
# 2 1 1 UA LGA IAH 227
# 3 1 1 AA JFK MIA 160
# Verbindung schließen
dbDisconnect(con)
# [1] TRUE As we can see, just a few lines of code are enough to view the contents of a database table. However, working with databases—especially via the R API—comes with a small challenge: SQL proficiency is required. While this isn’t necessarily a major hurdle, as SQL is an intuitive declarative language, it does present difficulties when dealing with large datasets. For simple queries like the one shown in the example, SQL is easy to understand. However, when datasets become too large to retrieve with a simple SELECT * FROM query, aggregations must be performed directly within the database. If these queries become complex, SQL can quickly turn into a real obstacle for data scientists.
At statworx , we frequently rely on databases to seamlessly integrate our predictive systems into our clients' data processes. However, you don’t have to be a SQL expert to work efficiently with databases in R. Several R packages help make database interactions safer, more stable, and easier to manage. Three packages are presented below that make working with databases safer, more stable and easier.
Managing Connections Efficiently with pool
When working with databases, technical considerations—such as connection management—are often critical. Managing connections dynamically, for example in a Shiny app, can be cumbersome. If not handled properly, it may even cause the app to crash, as some databases limit the number of simultaneous connections to 16 by default. To prevent this, connections must always be properly closed once they are no longer needed. In the example code above, closing the connection is done at the end.
To make connection management more stable, the pool package can be used.
The pool package creates a smart connection manager, known as an object pool. The advantage of this approach is that pool handles connection creation at the beginning of a session and efficiently manages them throughout, ensuring optimal utilization of database connections. A key benefit of pool is that its functionality closely mirrors the DBI package, making it easy to integrate into existing workflows. Let’s take a closer look at how this works with an example.
# Paket laden
library(pool)
# Pool-Objekt erzeugen
pool %
dbGetQuery("SELECT month, day, carrier, origin, dest, air_time
FROM flights LIMIT 3")
# month day carrier origin dest air_time
# 1 1 1 UA EWR IAH 227
# 2 1 1 UA LGA IAH 227
# 3 1 1 AA JFK MIA 160
# Verbindung schließen
poolClose(pool) As we can see, the syntax has hardly changed. The only difference is that we manage the connection pool using the dedicated functions dbPool() and poolClose(). The pool itself handles the connectors required for database queries. The diagram below illustrates this process schematically. In simple terms, the user sends a query to the pool. The pool then determines which connector to use to forward the query to the database and returns the result.
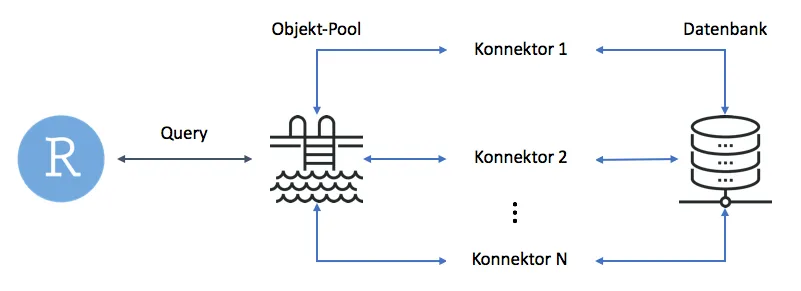
Hiding Credentials with config
When establishing a database connection, entering credentials is necessary, but they should be stored in a secure location. In the example above, this was not a concern because test_db was running locally on our computer, meaning the credentials were not sensitive. However, if the connection code needs to be shared with colleagues, it is best to retrieve credentials from an R object rather than hardcoding them directly.
With the config package, YAML configuration files can be read directly from R. These files can store database credentials and other sensitive settings in a human-readable format. To implement this, the first step is to create a configuration file named config.yml.
# Konfigurationsdatei erstellen
default:
database_settings:
host: 127.0.0.1
dbname: test_db
user: root
pwd: root
port: 3306
other_setting:
filepath: /path/to/file
username: gauss It is important to note that the first line of the file is mandatory. The YAML format offers the advantage of organizing settings thematically by introducing sub-lists. In the example, we created one sub-list containing database credentials (database_settings) and another for additional example settings (other_settings). In the next step, we can use the get() function to retrieve the database settings in a targeted manner.
# Paket laden
library(config)
# Datenbankeinstellungen laden
config <- get(value = "database_settings",
file = "~/Desktop/r-spotlight/config.yml")
str(config)
# List of 5
# $ host : chr "127.0.0.1"
# $ dbname: chr "test_db"
# $ user : chr "root"
# $ pwd : chr "root"
# $ port : int 3306
When creating a pool, we no longer have to disclose our sensitive data.
# Pool-Objekt erzeugen
pool <- dbPool(drv = RMySQL::MySQL(),
user = config$user,
password = config$pwd,
host = config$host,
port = config$port,
dbname = config$dbname)
SQL Without SQL Thanks to dbplyr
dbplyr serves as the database backend for dplyr, ensuring that dplyr’s elegant syntax can also be used when working with database connection objects. Since dbplyr is integrated into dplyr, there is no need to load it separately.
# Paket laden
library(dplyr)
# Eine kleine Query mit dplyr
pool %>%
tbl("flights") %>%
select(month, day, carrier, origin, dest, air_time) %>%
head(n = 3)
# Source: lazy query [?? x 6]
# Database: mysql 5.6.35 [root@127.0.0.1:/test_db]
# month day carrier origin dest air_time
#
# 1 1 1 UA EWR IAH 227
# 2 1 1 UA LGA IAH 227
# 3 1 1 AA JFK MIA 160
It is noticeable that the result is not an R data frame (as indicated by the "Source: lazy query ..." message). Instead, the R syntax is translated into SQL and sent as a query to the database. This means that all computations are performed directly on the database. The underlying SQL command can be displayed using the show_query() function.
# SQL anzeigen lassen
pool %>%
tbl("flights") %>%
select(month, day, carrier, origin, dest, air_time) %>%
head(n = 3) %>%
show_query()
# :SQL:
# SELECT `month` AS `month`, `day` AS `day`, `carrier` AS `carrier`,
# `origin` AS `origin`, `dest` AS `dest`, `air_time` AS `air_time`
# FROM `flights`
# LIMIT 3
Admittedly, this was not a complex query, but the principle should be clear. With this tool, it is easy to write much more sophisticated queries efficiently. For example, we could now calculate the average flight distance per airline directly within the database.
# Eine etwas komplexere Query
qry %
tbl("flights") %>%
group_by(carrier) %>%
summarise(avg_dist = mean(distance)) %>%
arrange(desc(avg_dist)) %>%
head(n = 3)
qry
# Source: lazy query [?? x 2]
# Database: mysql 5.6.35 [root@127.0.0.1:/test_db]
# Ordered by: desc(avg_dist)
# carrier avg_dist
#
# 1 HA 4983.000
# 2 VX 2499.482
# 3 AS 2402.000 We can use collect() to save the result of the SQL statement as an R object.# Save SQL result in R
# SQL Resultat in R abspeichern
qry %>%
collect()
# A tibble: 3 x 2
# carrier avg_dist
#
# 1 HA 4983.000
# 2 VX 2499.482
# 3 AS 2402.000
Conclusion
Working with databases can be made significantly easier and more secure by using the right packages. Thanks to dplyr, we no longer need to spend time memorizing SQL tutorials online just to write complex queries. Instead, we can focus on more important tasks.
References
- Boergs, Barbara (2017). Pool: Object Pooling. R Package Version 0.1.3. URL: https://CRAN.R-project.org/package=pool
- Datenbanken verstehen. Was ist eine Datenbank? URL: http://www.datenbanken-verstehen.de/datenbank-grundlagen/datenbank/
- Wickham, Hadley (2017a). Flights that Departed NYC in 2013. R Package. URL: https://CRAN.R-project.org/package=nycflights13
- Wickham, Hadley (2017b). dbplyr: A 'dplyr' Back End for Databases. R Package Version 1.1.0. URL: https://CRAN.R-project.org/package=dbplyr







